LaminDB is an open-source data lakehouse to enable learning at scale in biology. It organizes datasets through validation & annotation and provides data lineage, queryability, and reproducibility on top of FAIR data.
Why?
Reproducing analytical results or understanding how a dataset or model was created can be a pain. Let alone training models on historical data, LIMS & ELN systems, orthogonal assays, or datasets generated by other teams. Even maintaining a mere overview of a project's or team's datasets & analyses is harder than it sounds.
Biological datasets are typically managed with versioned storage systems, GUI-focused community or SaaS platforms, structureless data lakes, rigid data warehouses (SQL, monolithic arrays), and data lakehouses for tabular data.
LaminDB extends the lakehouse architecture to biological registries & datasets beyond tables (DataFrame, AnnData, .zarr, .tiledbsoma, ...) with enough structure to enable queries and enough freedom to keep the pace of R&D high.
Moreover, it provides context through data lineage -- tracing data and code, scientists and models -- and abstractions for biological domain knowledge and experimental metadata.
Highlights.
- data lineage: track inputs & outputs of notebooks, scripts, functions & pipelines with a single line of code
- unified infrastructure: access diverse storage locations (local, S3, GCP, ...), SQL databases (Postgres, SQLite) & ontologies
- lakehouse capabilities: manage, monitor & validate features, labels & dataset schemas; perform distributed queries and batch loading
- biological data formats: validate & annotate formats like
DataFrame,AnnData,MuData, ... backed byparquet,zarr, HDF5, LanceDB, DuckDB, ... - biological entities: organize experimental metadata & extensible ontologies in registries based on the Django ORM
- reproducible & auditable: auto-version & timestamp execution reports, source code & compute environments, attribute records to users
- zero lock-in & scalable: runs in your infrastructure; is not a client for a rate-limited REST API
- extendable: create custom plug-ins for your own applications based on the Django ecosystem
- integrations: visualization tools like vitessce, workflow managers like nextflow & redun, and other tools
- production-ready: used in BigPharma, BioTech, hospitals & top labs
LaminDB can be connected to LaminHub to serve as a LIMS for wetlab scientists, closing the drylab-wetlab feedback loop: lamin.ai
Copy summary.md into an LLM chat and let AI explain or read the docs.
Install the lamindb Python package:
pip install lamindbCreate a LaminDB instance:
lamin init --storage ./quickstart-data # or s3://my-bucket, gs://my-bucketOr if you have write access to an instance, connect to it:
lamin connect account/nameTrack a script or notebook run with source code, inputs, outputs, logs, and environment.
import lamindb as ln
ln.track() # track a run
open("sample.fasta", "w").write(">seq1\nACGT\n")
ln.Artifact("sample.fasta", key="sample.fasta").save() # create an artifact
ln.finish() # finish the runThis code snippet creates an artifact, which can store a dataset or model as a file or folder in various formats.
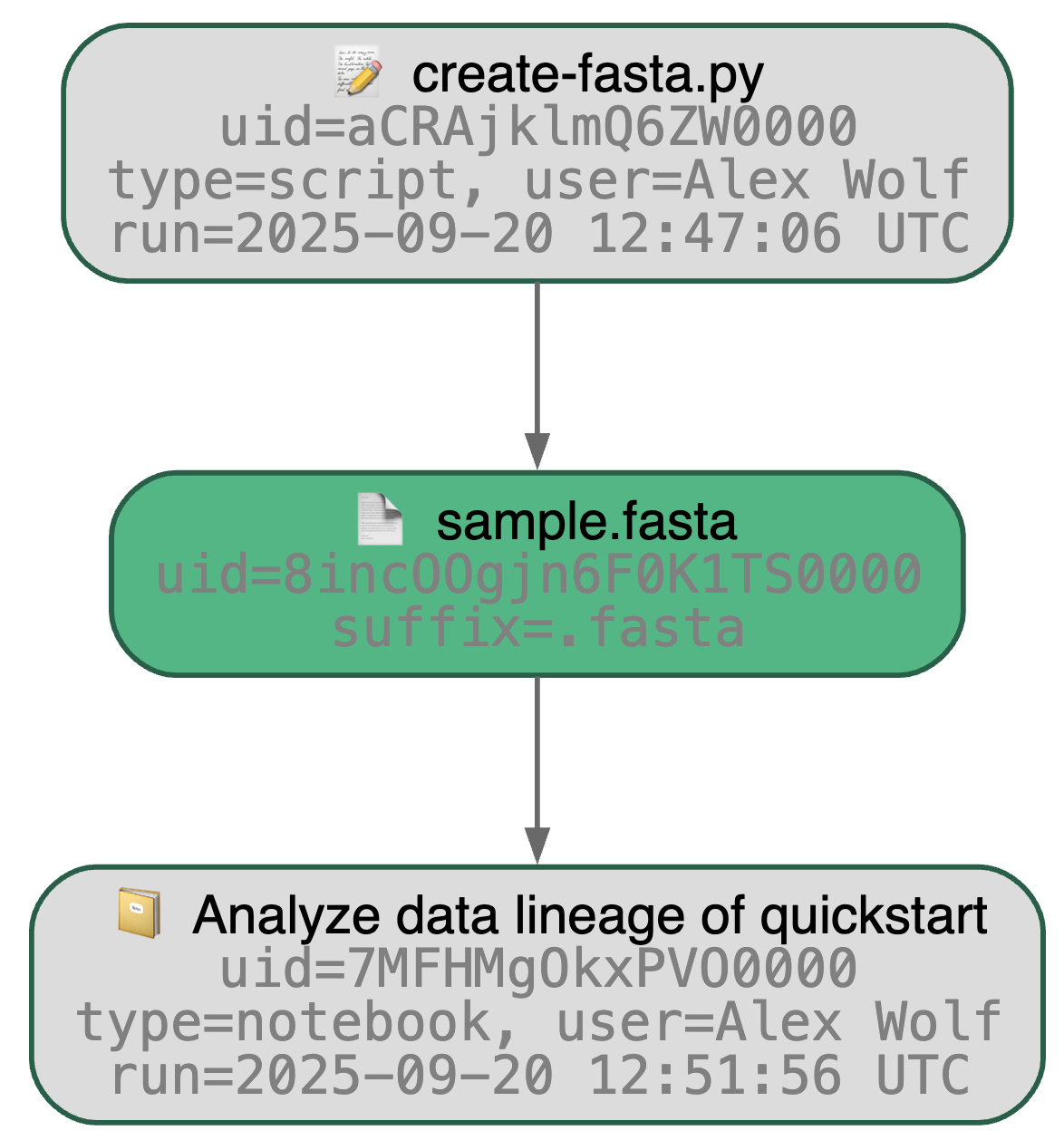
Running the snippet as a script (python create-fasta.py) produces the following data lineage.
artifact = ln.Artifact.get(key="sample.fasta") # query artifact by key
artifact.view_lineage()You'll know how that artifact was created and what it's used for (interactive visualization) in addition to capturing basic metadata:
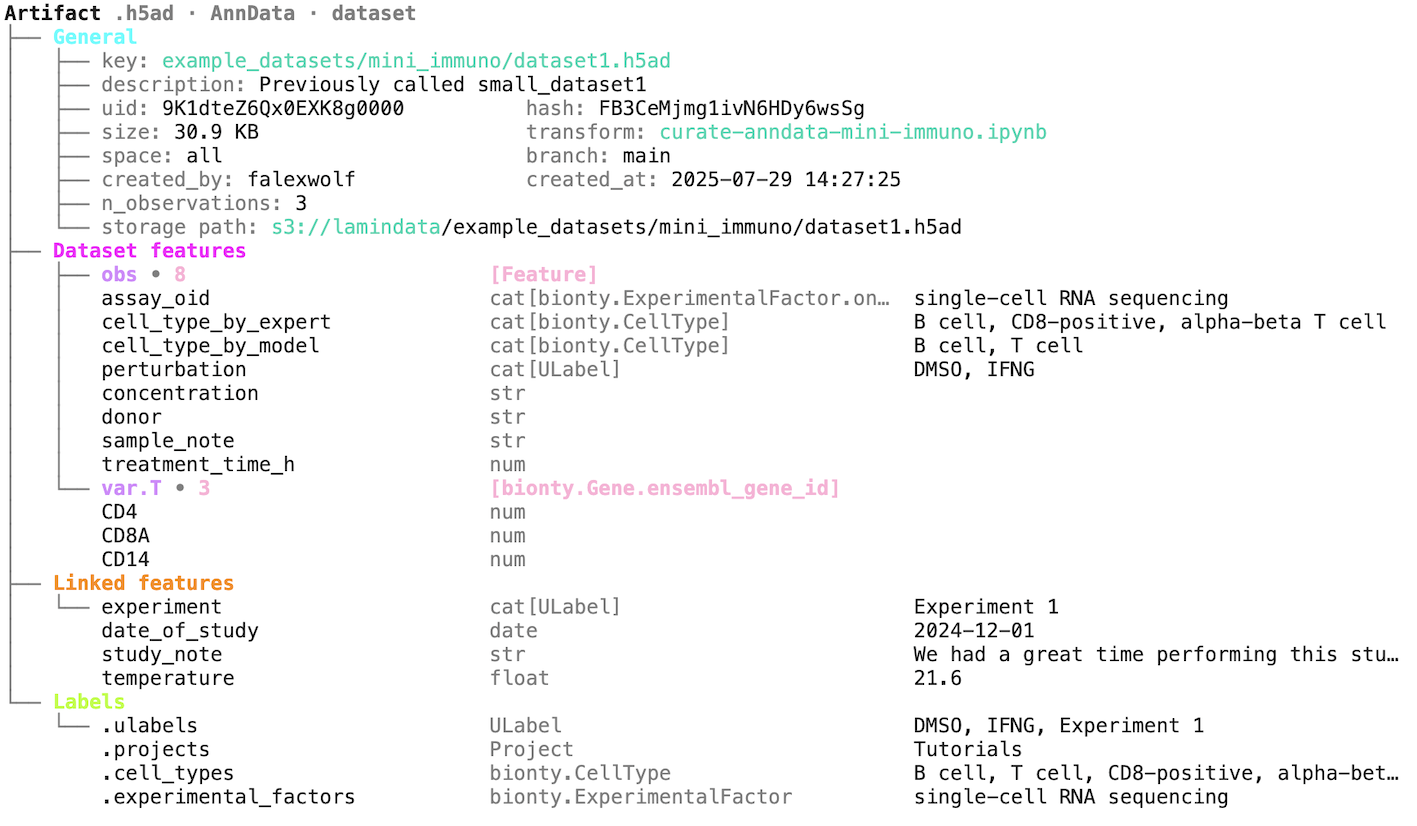
artifact.describe()You can organize datasets with validation & annotation of any kind of metadata to then access them via queries & search. Here is a more comprehensive example.
To annotate an artifact with a label, use:
my_experiment = ln.ULabel(name="My experiment").save() # create a label in the universal label ontology
artifact.ulabels.add(my_experiment) # annotate the artifact with the labelTo query for a set of artifacts, use the filter() statement.
ln.Artifact.filter(ulabels=my_experiment, suffix=".fasta").to_dataframe() # query by suffix and the ulabel we just created
ln.Artifact.filter(transform__key="create-fasta.py").to_dataframe() # query by the name of the script we just ranIf you have a structured dataset like a DataFrame, an AnnData, or another array, you can validate the content of the dataset (and parse annotations).
Here is an example for a dataframe: docs.lamin.ai/introduction#validate-an-artifact.
With a large body of validated datasets, you can then access data through distributed queries & batch streaming, see here: docs.lamin.ai/arrays.