This is a micro-package, containing the single class MultiVarGaussianKDE (and
helper function gaussian_kde) to estimate the probability density function of
a multivariate dataset using a Gaussian kernel. This package modifies the
jax.scipy.stats.gaussian_kde class (which is based on the
scipy.stats.gaussian_kde class), but allows for full control over the
covariance matrix of the kernel, even per-dimension bandwidths. See the
Documentation below for more information.
pip install mvgkdeFor these examples we will use the following imports:
import jax.numpy as jnp
import jax.random as jr
import matplotlib.pyplot as plt
import numpy as np
from mvgkde import MultiVariateGaussianKDE, gaussian_kde # This packageAnd we will generate a dataset to work with:
key = jr.key(0)
dataset = jr.normal(key, (2, 1000))Lastly we will define a plotting function:
# Create a grid of points
(xmin, ymin) = dataset.min(axis=1)
(xmax, ymax) = dataset.max(axis=1)
X, Y = np.mgrid[xmin:xmax:100j, ymin:ymax:100j]
positions = np.vstack([X.ravel(), Y.ravel()])
def plot_kde(kde: MultiVariateGaussianKDE) -> plt.Figure:
# Evaluate the KDE on the grid
Z = np.reshape(kde(positions).T, X.shape)
# Plot the results
fig, ax = plt.subplots()
ax.imshow(np.rot90(Z), cmap=plt.cm.gist_earth_r, extent=[xmin, xmax, ymin, ymax])
ax.plot(dataset[0], dataset[1], "k.", markersize=2)
ax.set(
title="2D Kernel Density Estimation using JAX",
xlabel="X-axis",
xlim=[xmin, xmax],
ylabel="Y-axis",
ylim=[ymin, ymax],
)
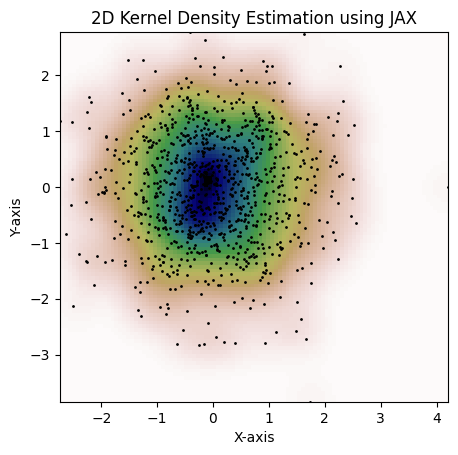
return figHere's an example that can be done with jax.scipy.stats.gaussian_kde:
kde = gaussian_kde(dataset, bw_method="scott")
fig = plot_kde(kde)
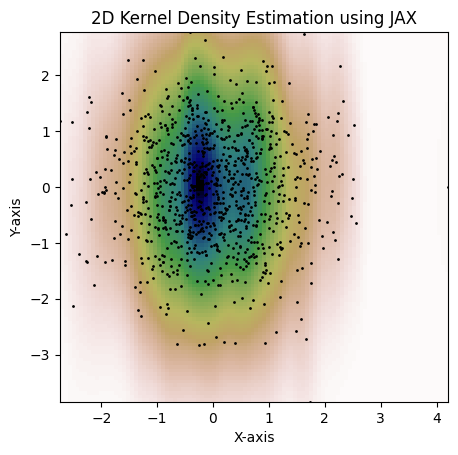
plt.show()Here's an example with a per-dimension bandwidth. This is not possible with the
jax.scipy.stats.gaussian_kde:
kde = gaussian_kde(dataset, bw_method=jnp.array([0.15, 1.3]))
fig = plot_kde(kde)
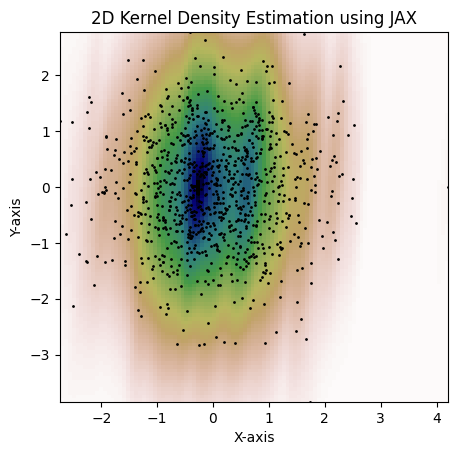
plt.show()Lastly, here's an example with 2D bandwidth matrix:
bw = jnp.array([[0.15, 3], [3, 1.3]])
kde = gaussian_kde(dataset, bw_method=bw)
fig = plot_kde(kde)
plt.show()The previous examples are using the convenience function gaussian_kde. This
actually just calls the constructor method
MultiVariateGaussianKDE.from_bandwidth. This function allows for customixing
the bandwidth factor on the data-driven covariance matrix, but does not allow
for specifying the covariance matrix directly. To do that, you can call the
MultiVariateGaussianKDE constructor directly, or the from_covariance
constructor method. To illustrate the difference between modifying the bandwidth
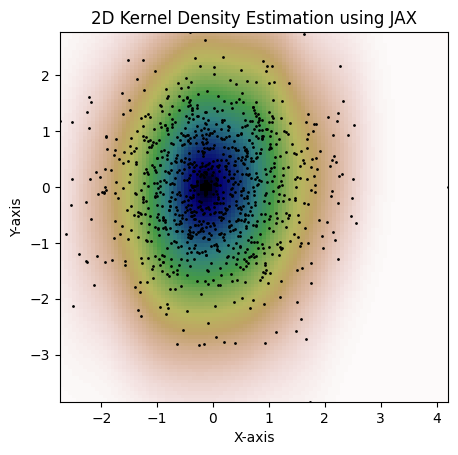
and setting the full covariance matrix, consider the following example:
kde = MultiVariateGaussianKDE.from_covariance(
dataset,
jnp.array([[0.15, 0.1], [0.1, 1.3]]),
)
fig = plot_kde(kde)
plt.show()This package modifies code from JAX, which is licensed under the Apache License 2.0.